Koide Laboratory - Technology
Our research is highly interdisciplinary, and we develop and utilize many technologies in:
- Directed evolution – phage display; yeast surface display
- Structure-guided design
- Biochemistry – protein technologies
- Biophysics – protein interactions; protein stability; protein structure
- Structural Biology – x-ray crystallography, nuclear magnetic resonance spectroscopy
- Recombinant DNA technologies – mutagenesis, protein expression
New technologies often catalyze major progress in science and engineering. The Koide group makes substantial investment of effort and resources to developing new technologies. We have pioneered a number of transformative technologies in protein design and engineering including the monobody and affinity clamp systems.
Monobody
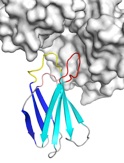 Monobodies are synthetic binding proteins that are constructed using a fibronectin type III domain (FN3) as a molecular scaffold. Monobodies are simple and robust alternative to antibodies in creating target-binding proteins. In 1998, we published the first paper demonstrating the monobody concept using the tenth FN3 domain of human fibronectin. Monobodies are produced from combinatorial libraries in which portions of the FN3 scaffold are diversified using highly tailored mixtures of amino acids by utilizing phage display and yeast surface display techniques. We have successfully generated a large number of monobodies that have high affinity and high specificity to their respective targets. Most monobodies bind a functional site of their targets and thus the monobody technology offers a powerful means to rapidly producing highly potent inhibitors.
Monobodies are synthetic binding proteins that are constructed using a fibronectin type III domain (FN3) as a molecular scaffold. Monobodies are simple and robust alternative to antibodies in creating target-binding proteins. In 1998, we published the first paper demonstrating the monobody concept using the tenth FN3 domain of human fibronectin. Monobodies are produced from combinatorial libraries in which portions of the FN3 scaffold are diversified using highly tailored mixtures of amino acids by utilizing phage display and yeast surface display techniques. We have successfully generated a large number of monobodies that have high affinity and high specificity to their respective targets. Most monobodies bind a functional site of their targets and thus the monobody technology offers a powerful means to rapidly producing highly potent inhibitors.
A major advantage of monobodies over conventional antibodies is that monobodies can readily be used as genetically encoded intracellular inhibitors, that is you can express a monobody inhibitor in a cell of choice by simply transfecting the cell with a monobody expression vector. This is because of unique characteristics of the underlying FN3 scaffold: small (~90 residues), stable, easy to produce, and its lack of disulfide bonds that makes it possible to produce functional monobodies in the reducing environment of the cytoplasm. In contrast, most antibodies and antibody fragments must be produced under oxidizing conditions, due to their dependence on disulfide bonds, and their two chains must be properly assembled.
Since our original publication, the FN3 scaffold has become the most widely adopted system for generating non-antibody binding proteins. The Koide group continues to push the Monobody technology forward and develop novel applications.
Monobody references.
Affinity Clamp
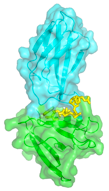 Affinity Clamps are synthetic proteins designed for recognizing short peptide motifs with high specificity and high affinity. The "Affinity Clamping" concept represents major advancement in protein design in that it combines two protein domains and evolving the domain interface for molecular recognition. Affinity Clamping mimics nonhomologous recombination in natural genetic evolution, and it is distinctly different from the concept underlying conventional binding protein design, i.e. alteration of a small patch within an existing protein scaffold.
Affinity Clamps are synthetic proteins designed for recognizing short peptide motifs with high specificity and high affinity. The "Affinity Clamping" concept represents major advancement in protein design in that it combines two protein domains and evolving the domain interface for molecular recognition. Affinity Clamping mimics nonhomologous recombination in natural genetic evolution, and it is distinctly different from the concept underlying conventional binding protein design, i.e. alteration of a small patch within an existing protein scaffold.
We have demonstrated that Affinity Clamping can achieve extraordinarily high specificity to a level sufficient for capturing a single peptide motif within a cell. By exploiting this transformative concept, we are systematically generating Affinity Clamps that recognize the C-terminal segments of human proteins (human "C-terminome") and phospho-tyrosine containing motifs involved in signal transduction and cancer biology.
Affinity Clamp references.
Synthetic Antibody
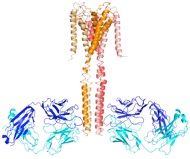 Antibodies remain essential tools in biology and medicine. In collaboration with Prof. Sachdev Sidhu (University of Toronto) and Tony Kossiakoff (University of Chicago), we have designed and constructed powerful antibody libraries. Our designs exploit a transformative discovery by the Sidhu group that only small subsets of the 20 amino acids, in particular tyrosine, are required for constructing a functional antigen-binding site. This concept makes it possible to design an antibody combinatorial library that is highly enriched in binding-competent sequences and thus to overcome the size limitation (~1E11, or 100 billions) of typical phage-display libraries.
Antibodies remain essential tools in biology and medicine. In collaboration with Prof. Sachdev Sidhu (University of Toronto) and Tony Kossiakoff (University of Chicago), we have designed and constructed powerful antibody libraries. Our designs exploit a transformative discovery by the Sidhu group that only small subsets of the 20 amino acids, in particular tyrosine, are required for constructing a functional antigen-binding site. This concept makes it possible to design an antibody combinatorial library that is highly enriched in binding-competent sequences and thus to overcome the size limitation (~1E11, or 100 billions) of typical phage-display libraries.
We call our antibodies "synthetic antibodies" to distinguish them from other antibodies derived from natural immune repertoires. Because we know precisely the design of our synthetic antibody libraries, analysis of synthetic antibodies enables us to iteratively improve our libraries. The current generation of our antibody library is sufficiently powerful to produce high affinity antibodies (dissociation constant in the low nanomolar range) to most structured proteins.
We have also developed a highly efficient method for selecting synthetic antibodies from these phage-display libraries. Importantly, because antibodies are selected in test tubes, not in animals as in the conventional immunization methods, we have a complete control over the conditions under which an antibody library is sorted. Furthermore, we can implement both positive and negative selection schemes, which ensures extremely high specificity of selected antibodies.
We have generated numerous synthetic antibodies to diverse types of targets including water-soluble proteins, detergent-solubilized membrane proteins and structured RNAs. These antibodies have proven to be transformative tools in structural biology, cell biology and molecular biology.
Synthetic Antibody references.
Molecular Display Technologies
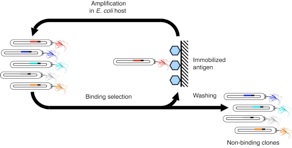 We extensively utilize the so-called molecular display technologies including phage-display and yeast surface display, which enables the establishment of the phenotype-genotype linkage at the level of protein function. Whereas we did not invent these technologies, we have developed numerous improvements to them from the necessity of adopting them to our projects – standard molecular display technology often fails when it is applied to a new protein system, for example the monobody system, and consequently we have gained expertise in display technologies. We often combine phage display and yeast display to accelerate synthetic protein generation.
We extensively utilize the so-called molecular display technologies including phage-display and yeast surface display, which enables the establishment of the phenotype-genotype linkage at the level of protein function. Whereas we did not invent these technologies, we have developed numerous improvements to them from the necessity of adopting them to our projects – standard molecular display technology often fails when it is applied to a new protein system, for example the monobody system, and consequently we have gained expertise in display technologies. We often combine phage display and yeast display to accelerate synthetic protein generation.
Molecular Display references.
Antibody Validation
As we develop novel binding proteins, we often need to characterize existing reagents that bind to the same target. This has led us to develop a new method for quantitatively characterizing the affinity and specificity of polyclonal and monoclonal antibodies. Using this method we analyze “ChIP grade” antibodies to histone posttranslational modifications. Sadly we have found many of these commercial antibodies are nowhere near “ChIP grade”.
Antibody Validation page.